|
Mitochondrial biogenesis in Trypanosoma brucei
Mitochondria do not form de novo but derive from preexisting organelles by processes collectively referred to as mitochondrial biogenesis. These include import of most mitochondrial proteins and, in most eukaryotes, import of at least some tRNAs. Furthermore, a limited set of proteins that is essential for oxidative phosphorylation need to be synthesized inside mitochondria.
Our research group is working on mitochondrial biogenesis using the parasitic protozoa Trypanosoma brucei as a model. T. brucei and its relatives belong to the earliest diverging branches of the eukaryotic evolutionary tree, which have bona fide mitochondria involved in oxidative phosphorylation. Their mitochondria therefore show many peculiarities. The most important ones in the context of our studies are a complete lack of mitochondrial tRNA genes that is compensated by import of cytosolic tRNAs and an apparently minimized mitochondrial protein import system. Moreover, T. brucei is the causative agent of human sleeping sickness and therefore of great clinical importance.
Why should research of exotic trypanosome-specific features be of interest for basic science in general? The reason is that these superficially trypanosome-specific features are not really unique, but rather represent extreme examples of processes that in a more cryptic way occur in other cells as well. Thus, most eukaryotes import mitochondrial tRNAs, just not the entire set as T. brucei. All eukaryotes import most of their mitochondrial proteins, however, their import machineries are not as minimized as the one in T. brucei. Processes such as trans-splicing, glycosyl-phosphatidyl-inositol protein anchoring and RNA editing were originally discovered in T. brucei and at first regarded as exotic oddities of trypanosomes. Later on, however, they were shown to be widespread within eukaryotes and are now described in every cell biology textbook. Thus, T. brucei is an attractive experimentally highly tractable system to study basic as yet poorly understood biological processes.
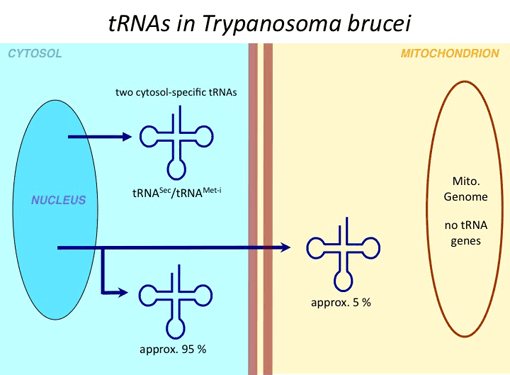
Mitochondrial tRNA import
The complete sequences of mitochondrial genomes are available for thousands of different species. Bioinformatic analysis of these sequences allows to predict the number of mitochondrial tRNA genes and to match them to the codons that are used by the mitochondrial translation systems. The conclusion of this analysis is that most eukaryotes lack a variable number of apparently essential mitochondrial tRNA genes that is compensated for by import of the corresponding cytosolic tRNAs. Mitochondrial tRNA import is very widespread, it occurs in most protozoa, in all plants, in many fungi and in a few animals. However, many eukaryotes including essentially all vertebrates whose mitochondrial genomes encode an apparently complete set of tRNAs are not expected to import tRNAs (for an update on this click here). In most organisms which import tRNAs a subset of mitochondrial tRNA genes has been retained. Thus, the complete lack of mitochondrial tRNA genes as seen in trypanosomatids such as T. brucei or Leishmania is highly unusual. The only other known taxonomic group where the same situation is found are the apicomplexans. Interestingly, the two groups are not closely related phylogenetically but share a parasitic lifestyle.
In all organisms studied so far imported nucleus-encoded mitochondrial tRNAs represent a small fraction of normal cytosolic tRNAs (note that nucleus-encoded tRNAs that are mitochondria specific do not exist). As a consequence imported tRNAs, contrary to the bacterial-type mitochondrially encoded tRNAs, are universally of the eukaryotic type. Interestingly, however, they have to function in the context of the bacterial-type translation system of mitochondria.
The three main questions regarding mitochondrial tRNA import that are investigated in our lab are:
- What determines the specificity and extent of the process?
- What is the machinery and the mechanism for membrane translocation of tRNAs?
- How are the imported tRNAs used in mitochondrial translation?
Mitochondrial protein import
The mitochondrial genome codes for only a small number of proteins so that almost all of the ~1000 proteins that constitute a mitochondrion are synthesized on cytosolic ribosomes and targeted to mitochondria post-translationally. Extensive studies during the last decades have identified four hetero-oligomeric protein complexes that catalyze mitochondrial protein import and whose core components are conserved among all eukaryotes. Two of these complexes are found in the outer membrane (the TOM complex and the SAM complex) and two in the inner membrane (the TIM22 complex and the TIM23 complex). A genome analysis revealed two aspects where the protein import pathway of T. brucei appears to be different and possibly simpler than in other eukaryotes. These are the apparent absence of a conventional TOM complex and the existence of only a single TIM complex. Thus, studying mitochondrial protein import in T. brucei is likely to provide more insight into the enigmatic evolutionary origin of mitochondrial protein import in general.
Further readings (Reviews only)
Salinas, T., Duchêne, A.M., and Maréchal-Drouard, L. (2008)
Recent advances in tRNA mitochondrial import
Trends Biochem. Sci. 33, 320-329.
Schneider, A., Bursać, D., and Lithgow, T. (2008)
The direct route: a simplified pathway for protein import into the mitochondrion of trypanosomes
Trends Cell Biol. 18, 12-18.
Tarassov, I., Kamenski, P., Kolesnikova, O., Karicheva, O., Martin, R.P., Krasheninnikov, I.A., and Entelis, N. (2007)
Import of nuclear DNA-encoded RNAs into mitochondria and mitochondrial translation
Cell Cycle 6, 2473-2477.
Schneider, A. (2001)
Unique aspects of mitochondrial biogenesis in trypanosomatids
Int. J. Parasitol. 31, 1403-1415.
Entelis, N.S., Kolesnikova, O.A., Martin, R.P., and Tarassov, I.A. (2001)
RNA delivery into mitochondria
Adv. Drug Deliv. Rev. 49, 199-215.
Schneider, A., and Marechal-Drouard, L. (2000)
Mitochondrial tRNA import: are there distinct mechanisms?
Trends Cell Biol. 10, 509-513.
Schneider, A. (1994)
Import of RNA into mitochondria
Trends Cell Biol. 4, 282-286. |
 |