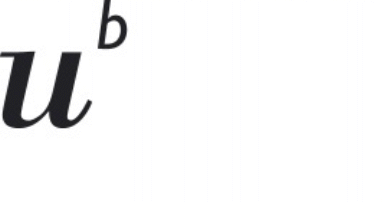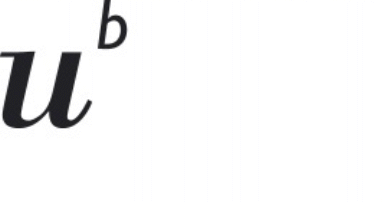|
Trypanosoma brucei as an experimental model
T. brucei is not only a clinically important pathogen but also an excellent system for basic research for the following practical reasons:
- The T. brucei genome has been sequenced (GeneDB) (http://www.genedb.org/genedb/tryp/) and excellent tools for reverse genetics are available. Linear DNA is integrated into the T. brucei genome by homologous recombination allowing the production of gene-knockouts. A very tight system for conditional expression of gene products is available. Furthermore, T. brucei was one of the first organisms where RNAi methods to inactivate specific gene products have been used on a routine basis. The use of inducible gene expression allows conditional RNAi, which is an excellent and fast method to analyze the function of both essential and non-essential gene products.
- T. brucei can be grown in quantities large enough for biochemical fractionations. A variety of protocols have been established which allow the purification of defined organelles including the nucleus, mitochondria, glycosomes and flagella.
- Despite the fact that T. brucei is a unicellular organism, it is a highly structured cell, large enough for sophisticated cell biological analyses such as high resolution immunofluorescence as well as electron microscopy.
Thus, using trypanosomes as an experimental model it is possible to combine genetic, biochemical and cell biological approaches. My research group is trying to use these advantages to study the mitochondrial biogenesis of T. brucei.
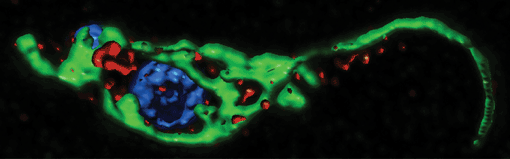
3D-reconstruction of a T. brucei cell from optical sections obtained by confocal microscopy. DNA is shown in blue and the mitochondrion in green. Red shows the localization of the dynamin-like protein that is required for mitochondrial fission and endocytosis. (Bild: Adrian Hehl)
|
 |